Entry information : OsPrx09(LOC_Os01g18950)
| Entry ID | 1019 |
|---|---|
| Creation | 2009-01-08 (Christophe Dunand) |
| Last sequence changes | 2011-02-15 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2022-02-09 (Christophe Dunand) |
Peroxidase information: OsPrx09(LOC_Os01g18950)
| Name | OsPrx09(LOC_Os01g18950) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx337] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Viridiplantae (green plants); Streptophyta; Angiospermae; Monocotyledons | ||||||||||||||||||||||||||||||||||||
| Organism | Oryza sativa ssp japonica cv Nipponbare [TaxId: 39947 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
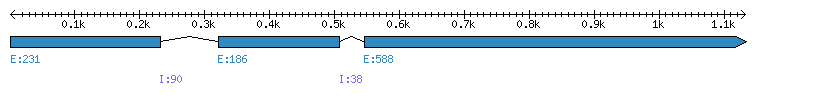
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references OsPrx09(LOC_Os01g18950)
| Literature | Passardi F., Longet, D., Penel C. and Dunand, C. The class III peroxidase multigenic family in rice and its evolution in land plants Phytochemistry, 65 (13), 1879-1893 (2004) |
|---|---|
| Protein ref. | UniProtKB: Q5U1T4 |
| DNA ref. | GenBank: NC_008394.1 (10712323..10711218) |
Protein sequence: OsPrx09(LOC_Os01g18950)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 1, intron 1 and 2). No EST. Missing 'c'. Incorrect prediction from Phytozome (splicing error exon 3 | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
