Entry information : CbraMSD02 (g360/ CHBRA09g00790)
| Entry ID | 13795 |
|---|---|
| Creation | 2016-06-21 (Christophe Dunand) |
| Last sequence changes | 2016-06-21 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2016-10-20 (Christophe Dunand) |
Peroxidase information: CbraMSD02 (g360/ CHBRA09g00790)
| Name (synonym) | CbraMSD02 (g360/ CHBRA09g00790) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Mn SOD [Orthogroup: MSD002] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Charophyceae Characeae Chara | ||||||||||||||||||||||||||||||||||||
| Organism | Chara braunii [TaxId: 69332 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
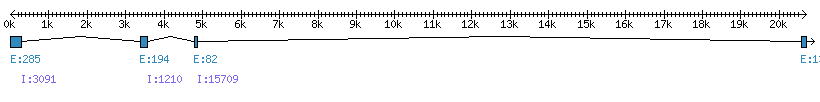
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references CbraMSD02 (g360/ CHBRA09g00790)
Protein sequence: CbraMSD02 (g360/ CHBRA09g00790)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | |||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
