Entry information : EsilMDAR01 (Esi_0039_0037)
| Entry ID | 16938 |
|---|---|
| Creation | 2021-01-30 (Christophe Dunand) |
| Last sequence changes | 2021-01-30 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
Peroxidase information: EsilMDAR01 (Esi_0039_0037)
| Name (synonym) | EsilMDAR01 (Esi_0039_0037) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Monodhydroascorbate Reductase [Orthogroup: MDAR002] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Phaeophyceae Ectocarpaceae Ectocarpus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Ectocarpus siliculosus [TaxId: 2880 ] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
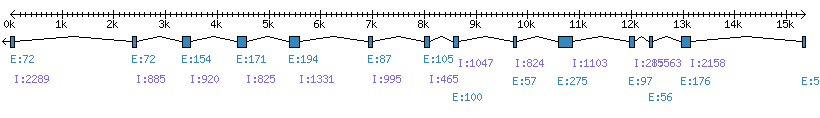
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Literature and cross-references EsilMDAR01 (Esi_0039_0037)
| Protein ref. | GenBank: CBJ48589.1 |
|---|---|
| DNA ref. | GenBank: FN648586.1 (195429..180069) |
Protein sequence: EsilMDAR01 (Esi_0039_0037)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | |||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
