Entry information : ZmPrx03(Zm00001d037547 / GRMZM2G135 / pox3108 [6a])
| Entry ID | 19 |
|---|---|
| Creation | 2006-08-28 (Christophe Dunand) |
| Last sequence changes | 2011-01-20 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Christophe Dunand |
| Last annotation changes | 2023-01-09 (Christophe Dunand) |
Peroxidase information: ZmPrx03(Zm00001d037547 / GRMZM2G135 / pox3108 [6a])
| Name | ZmPrx03(Zm00001d037547 / GRMZM2G135 / pox3108 [6a]) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx093] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Monocotyledons Poaceae Zea | ||||||||||||||||||||||||||||||||||||
| Organism | Zea mays [TaxId: 4577 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
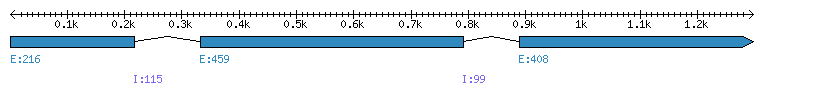
| Gene structure Fichier |
Exons►
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references ZmPrx03(Zm00001d037547 / GRMZM2G135 / pox3108 [6a])
| Literature | REFERENCE 1 de Obeso,M. et al., Gene 309 (1), 23-33 (2003).
REFERENCE 2 Guillet-Claude,C., Birolleau-Touchard,C., Manicacci,D., Rogowsky,P.M., Rigau,J., Murigneux,A., Martinant,J.P. and Barriere,Y. Nucleotide diversity of the ZmPox3 maize peroxidase gene: relationships between a MITE insertion in exon 2 and variation in forage maize digestibility. BMC Genet. 5 (1), 19 (2004) |
|---|---|
| Protein ref. | UniProtKB: Q6RFK3 |
| mRNA ref. | GenBank: AY500792 |
| Cluster/Prediction ref. | UniGene: Zm.2766 |
Protein sequence: ZmPrx03(Zm00001d037547 / GRMZM2G135 / pox3108 [6a])
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 6, intron 1 and 3') and mRNA. relationships with variation in cell wall digestibility. Containing IntR (3'). Also present in different cultivars with a MITE seq in the exon 2: B73 (AY500800), EP1 (AY500797), F120 (AY500809), F7 (AY508516), F226 (AY508161), F227 (AY508162), F271 (AY500812), F286 (AY500796), F324 (AY508163), F564 (AY500792), F64 (AY500794), F7012 (AY508160), F7025 (AY500804), MBS847 (AY500805), Quebec28 (AY500781, AY508159), W117 (AY500808), W64A (AY500789), GeneChip® Maize Genome Array ID: Zm.2766.1.A1_at |
||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
