Entry information : SaPrx02
| Entry ID | 2122 |
|---|---|
| Creation | 2007-01-12 (Christophe Dunand) |
| Last sequence changes | 2007-01-12 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2019-11-27 (Christophe Dunand) |
Peroxidase information: SaPrx02
| Name | SaPrx02 | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx423] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Orobanchaceae Striga | ||||||||||||||||||||||||||||||||||||
| Organism | Striga asiatica (witchweed) [TaxId: 4170 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | Seedlings |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
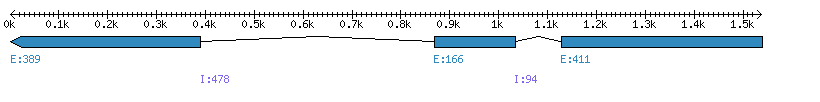
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references SaPrx02
| Literature | Kim,D., Kocz,R., Boone,L., Keyes,W.J. and Lynn,D.G. On becoming a parasite: evaluating the role of wall oxidases in parasitic plant development Chem. Biol. 5 (2), 103-117 (1998) |
|---|---|
| Protein ref. | GenBank: GER29790.1 UniProtKB: O49193 |
| mRNA ref. | GenBank: AF043235 [Other strain] |
Protein sequence: SaPrx02
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic and mRNA (different strain). | ||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
