Entry information : SbPrx83 (Sobic.005G011200.1 [2.1] / SB05G001000 [1.4])
| Entry ID | 2760 |
|---|---|
| Creation | 2008-10-12 (Christophe Dunand) |
| Last sequence changes | 2014-01-08 (Qiang Li) |
| Sequence status | complete |
| Reviewer | Messaoudi |
| Last annotation changes | 2014-08-04 (Messaoudi) |
Peroxidase information: SbPrx83 (Sobic.005G011200.1 [2.1] / SB05G001000 [1.4])
| Name (synonym) | SbPrx83 (Sobic.005G011200.1 [2.1] / SB05G001000 [1.4]) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx006] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Monocotyledons Poaceae Sorghum | ||||||||||||||||||||||||||||||||||||
| Organism | Sorghum bicolor [TaxId: 4558 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
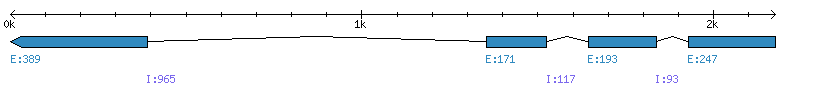
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references SbPrx83 (Sobic.005G011200.1 [2.1] / SB05G001000 [1.4])
| Literature | Plant genome Database |
|---|---|
| DNA ref. | Phytozome 12: chromosome_5 (1001461..998634) |
| Cluster/Prediction ref. | PlantGDB: SbGSStuc11-12-04.6734.1 |
Protein sequence: SbPrx83 (Sobic.005G011200.1 [2.1] / SB05G001000 [1.4])
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 5, 4 introns => extra introns between GRRD and VALS). No EST. In DNA sequence there is overlapping seq due to the bad assembly. The dna seq below is the DNA reassemblied. The extra intron is not really existing. Containing intron shift. | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
