Entry information : OsGPx04(LOC_Os06g08670)
| Entry ID | 2870 |
|---|---|
| Creation | 2007-08-15 (Christophe Dunand) |
| Last sequence changes | 2007-08-15 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2022-02-18 (Catherine ) |
Peroxidase information: OsGPx04(LOC_Os06g08670)
| Name | OsGPx04(LOC_Os06g08670) | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Plant glutathione peroxidase [Orthogroup: Gpx2001] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Viridiplantae (green plants); Streptophyta; Angiospermae; Monocotyledons | ||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Oryza sativa ssp japonica cv Nipponbare [TaxId: 39947 ] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||
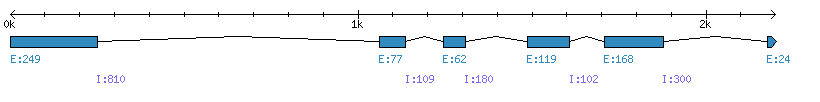
| Gene structure Fichier |
Exons►
|
||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence: OsGPx04(LOC_Os06g08670)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic and 38 ESTs (TC from TGI) and 46 EST (UniGene Os.7552 which compile japonica and indica together). Two TREMBL accessions: Q5SMW6 and A3B930 both contain extra sequences not confirmed by ESTs (splicing prediction error). Predicted sequence in Phytozome (LOC_Os06g08670) erroneous in C-term (prediction error for the last exon) |
||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
