Entry information : CsaPrx09 ( Cucsa.247030.1)
| Entry ID | 2919 |
|---|---|
| Creation | 2006-03-23 (Christophe Dunand) |
| Last sequence changes | 2016-02-04 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Achraf Jemmat |
| Last annotation changes | 2016-02-04 (Achraf Jemmat) |
Peroxidase information: CsaPrx09 ( Cucsa.247030.1)
| Name (synonym) | CsaPrx09 ( Cucsa.247030.1) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx030] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Cucurbitaceae Cucumis | ||||||||||||||||||||||||||||||||||||
| Organism | Cucumis sativus [TaxId: 3659 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | Flower buds |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
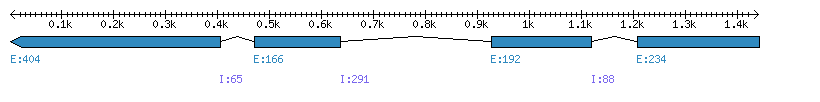
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references CsaPrx09 ( Cucsa.247030.1)
| Literature | Plant Genome Network. |
|---|---|
| DNA ref. | Phytozome 12: scaffold02188 (70759..69320) |
| EST ref. | GenBank: CV005121.1 [3' end] |
Protein sequence: CsaPrx09 ( Cucsa.247030.1)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic and 1 EST (CV005121). Incorrect prediction from Phytozome (5' end is missing). FHDCF instead of FHDCFVN due to a point mutation GTTAATGA instaed of GTTAATGC found in the other member of the orthogroup. | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
