Entry information : PpaPrx13 (Prx34a / Prx34b / PP00219G00030)
| Entry ID | 3419 |
|---|---|
| Creation | 2006-07-26 (Christophe Dunand) |
| Last sequence changes | 2012-01-05 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2011-01-25 (Qiang Li) |
Peroxidase information: PpaPrx13 (Prx34a / Prx34b / PP00219G00030)
| Name (synonym) | PpaPrx13 (Prx34a / Prx34b / PP00219G00030) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx063] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Funariaceae Physcomitrella | ||||||||||||||||||||||||||||||||||||
| Organism | Physcomitrella patens [TaxId: 3218 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducers | Elicitor treatment (glucan, oligosaccharides, yeast extract, Phytophthora crude extract) Pathogen interaction |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
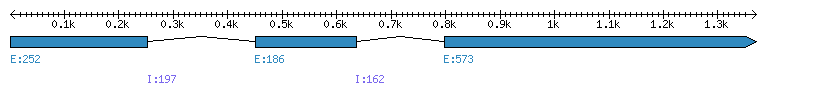
| Gene structure Fichier |
Exons►
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references PpaPrx13 (Prx34a / Prx34b / PP00219G00030)
| Literature | REFERENCE 1 Fujita,T., Nishiyama,T., Shin-i,T., Kohara,Y. and Hasebe,M. Physcomitrella patens EST at a stage of the first asymmetric cell division of protoplasts.
REFERENCE 2 Lehtonen MT, Akita M, Kalkkinen N, et al. Quickly-released peroxidase of moss in defense against fungal invaders. NEW PHYTOLOGIST, 183, (2), 432-443 |
|---|---|
| DNA ref. | Phytozome 12: scaffold_219 (45619..46988) |
| Cluster/Prediction ref. | UniGene: Ppa.7622 |
Protein sequence: PpaPrx13 (Prx34a / Prx34b / PP00219G00030)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (introns 1 and 2) and 12 ESTs. Inverse tandem duplication or assembly error. Incorrect prediction from Phytozome (PS is missing). | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
