Entry information : Os1CysPrx01(LOC_Os07g44430 / RAB24)
| Entry ID | 4023 |
|---|---|
| Creation | 2006-11-03 (Nicolas Rouhier) |
| Last sequence changes | 2006-11-03 (Nicolas Rouhier) |
| Sequence status | complete |
| Reviewer | Christophe Dunand |
| Last annotation changes | 2022-02-13 (Christophe Dunand) |
Peroxidase information: Os1CysPrx01(LOC_Os07g44430 / RAB24)
| Name | Os1CysPrx01(LOC_Os07g44430 / RAB24) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | 1-Cysteine peroxiredoxin [Orthogroup: 1CysPrx001] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Viridiplantae (green plants); Streptophyta; Angiospermae; Monocotyledons | ||||||||||||||||||||||||||||||||||||
| Organism | Oryza sativa ssp japonica cv Nipponbare [TaxId: 39947 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue types | Callus Flowers Inflorescences Mixed tissues Seeds Stems Whole plant |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
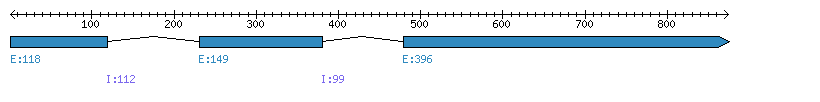
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references Os1CysPrx01(LOC_Os07g44430 / RAB24)
| Literature | Gama et physiologia plantarum in press;
Dietz et al The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot. 2006;57(8):1697-709. Yao Q., Peng R., Xiong A. Identification a novel antioxidant enzyme from rice. |
|---|---|
| Protein ref. | UniProtKB: P0C5C9 UniProtKB_Uniparc: A3BMM5 Q0D4A2 |
| DNA ref. | GenBank: NC_008400.1 (26495351..26496221) |
| Cluster/Prediction ref. | UniGene: Os.13972 |
Protein sequence: Os1CysPrx01(LOC_Os07g44430 / RAB24)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 7, 2 introns), 6 cDNA and 299 ESTs. Cultivarv="Japonica". Three other SwissProt/TREMBL accessions. Incorrect prediction from Phytozome (splicing error, extra exon/intron on 3') | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
