Entry information : Ani1CysPrx01
| Entry ID | 4961 |
|---|---|
| Creation | 2007-03-13 (Filippo Passardi) |
| Last sequence changes | 2012-04-25 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2012-04-25 (Christophe Dunand) |
Peroxidase information: Ani1CysPrx01
| Name | Ani1CysPrx01 | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | 1-Cysteine peroxiredoxin [Orthogroup: 1CysPrx001] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Fungi Ascomycota Eurotiomycetes Trichocomaceae Emericella | ||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Aspergillus nidulans-Emericella nidulans [TaxId: 162425 ] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||
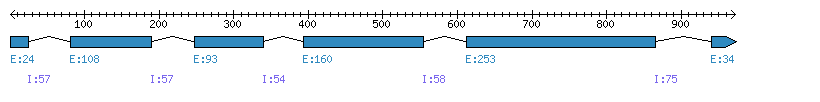
| Gene structure Fichier |
Exons►
|
||||||||||||||||||||||||||||||||||||||||||||||||
Literature and cross-references Ani1CysPrx01
| Literature | Galagan J.E. et al. Sequencing of Aspergillus nidulans and comparative analysis with A.fumigatus and A. oryzae. Nature 438 (7071), 1105-1115 (2005)
|
|---|---|
| Protein ref. | GenPept: EAA64812 [Incorrect prediction] UniProtKB: Q5BCN8 [Incorrect prediction] |
| DNA ref. | JGI genome: ChrVII_A_nidulans_FGSC_A4 (1631541..1632513) |
| mRNA ref. | GenBank: EAA64812 [Incorrect prediction] |
Protein sequence: Ani1CysPrx01
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from Genomic (Chromo VII, 6 exons). No EST (EST found with other aspergillus). Incorrect prediction in NCBI/SwissProt (too long, splicing is incorrect --> checked with FGeneSH-SoftBerry) and JGI (last short exon is missing). Strain="FGSC A4". |
||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
