Entry information : AfumDyPrx03
| Entry ID | 5102 |
|---|---|
| Creation | 2007-10-22 (Christophe Dunand) |
| Last sequence changes | 2012-01-26 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2012-01-26 (Christophe Dunand) |
Peroxidase information: AfumDyPrx03
| Name | AfumDyPrx03 | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | DyP-type peroxidase D [Orthogroup: DyPrxD001] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Fungi Ascomycota Eurotiomycetes Trichocomaceae Neosartorya | ||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Aspergillus fumigatus [TaxId: 746128 ] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
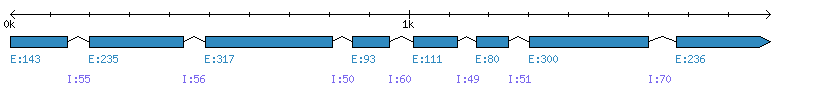
|
||||||||||||||||||||||||||||||||||||||||||||||||
Literature and cross-references AfumDyPrx03
| Literature | Nierman W.C. et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156(2005). |
|---|---|
| Protein ref. | GenPept: EAL86784 UniProtKB: Q4WH07 |
| DNA ref. | GenBank: AAHF01000009 (377028..378933) |
Protein sequence: AfumDyPrx03
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from Genomic (chromo 7, 8 exons). Strain="Af293 / CBS 101355 / FGSC A1100". Sequence was corrected from genomic NCBI data (START was predicted a little too early) and SwissProt data (large deletion wrongly predicted) with SoftBerry (FGeneSH). | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
