Entry information : ZmAPx02(Zm00001d007234 / GRMZM2G140667 [6a] / APx2)
| Entry ID | 6705 |
|---|---|
| Creation | 2009-04-12 (Christophe Dunand) |
| Last sequence changes | 2011-01-20 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2023-01-19 (Christophe Dunand) |
Peroxidase information: ZmAPx02(Zm00001d007234 / GRMZM2G140667 [6a] / APx2)
| Name | ZmAPx02(Zm00001d007234 / GRMZM2G140667 [6a] / APx2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Ascorbate peroxidase [Orthogroup: APx2001] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Monocotyledons Poaceae Zea | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Zea mays [TaxId: 4577 ] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
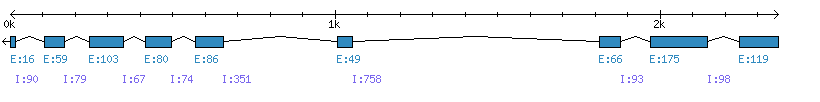
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence: ZmAPx02(Zm00001d007234 / GRMZM2G140667 [6a] / APx2)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 2, 8 introns), 6 mRNA sequences and 145 EST sequences. Incorrect prediction from Phytozome (extra aa around the splicing sites). Strain="B73" and "hybrid 35A19". | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
