Entry information : ZmPrx29(Zm00001d006097 / Peroxidase 73)
| Entry ID | 725 |
|---|---|
| Creation | 2005-12-15 (Christophe Dunand) |
| Last sequence changes | 2005-12-15 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Christophe Dunand |
| Last annotation changes | 2022-12-31 (Christophe Dunand) |
Peroxidase information: ZmPrx29(Zm00001d006097 / Peroxidase 73)
| Name | ZmPrx29(Zm00001d006097 / Peroxidase 73) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx031] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Monocotyledons Poaceae Zea | ||||||||||||||||||||||||||||||||||||
| Organism | Zea mays [TaxId: 4577 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue types | Ears Seedlings Silks |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
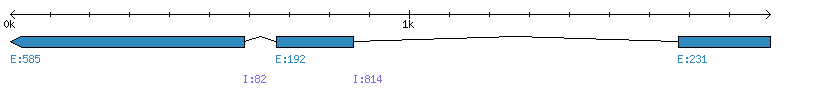
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references ZmPrx29(Zm00001d006097 / Peroxidase 73)
| Literature | Gardiner,J., Schroeder,S., Polacco,M.L., Sanchez-Villeda,H., Fang,Z., Morgante,M., Landewe,T., Fengler,K., Useche,F., Hanafey,M., Tingey,S., Chou,H., Wing,R., Soderlund,C. and Coe,E.H. Jr. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 134 (4), 1317-1326 (2004). |
|---|---|
| Protein ref. | UniProtKB: B4F8B7 B6U0T8 |
| DNA ref. | GenBank: AC200703.3 (64112..66012) |
| mRNA ref. | GenBank: AY111875.1 |
| Cluster/Prediction ref. | UniGene: Zm.1415 |
Protein sequence: ZmPrx29(Zm00001d006097 / Peroxidase 73)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 2, introns 1 and 2) and 35 ESTs. Cultivar="F2", "B73". | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
