Entry information : HcyPrxQ
| Entry ID | 7611 |
|---|---|
| Creation | 2010-09-21 (Christophe Dunand) |
| Last sequence changes | 2012-04-03 (Catherine Mathe (Scipio)) |
| Sequence status | complete |
| Reviewer | Achraf Jemmat |
| Last annotation changes | 2015-12-22 (Achraf Jemmat) |
Peroxidase information: HcyPrxQ
| Name | HcyPrxQ | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Atypical 2-Cysteine peroxiredoxin (type Q) [Orthogroup: PrxQ001] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Fungi Basidiomycota Agaricomycetes Cortinariaceae Hebeloma | ||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Hebeloma cylindrosporum [TaxId: 76867 ] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
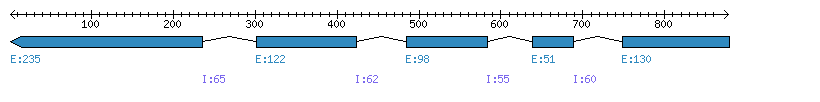
|
||||||||||||||||||||||||||||||||||||||||||||||||
Literature and cross-references HcyPrxQ
| Literature | Lambilliotte,R., Cooke,R., Samson,D., Fizames,C., Gaymard,F., Plassard,C., Tatry,M.V., Berger,C., Laudie,M., Legeai,F., Karsenty ,E., Delseny,M., Zimmermann,S. and Sentenac,H. Large scale identification of genes in the fungus Hebeloma cylindrosporum paves the way to molecular analyses of ectomycorrhizal symbiosis. New Phytol 164 (3), 505-513 (2004). |
|---|---|
| DNA ref. | JGI genome: scaffold_9 (496849..495972) |
| EST ref. | GenBank: CK993011.1 [5' end] CK995357.1 [5' end] |
Protein sequence: HcyPrxQ
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from 2 ESTs. 3'end is largely divergente from other orthologs(!!!) | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
