Entry information : ZmPrx70(Zm00001d022458 / GRMZM2G025441 / pmPOX2b / PRX70 / Peroxidase 70)
| Entry ID | 780 |
|---|---|
| Creation | 2009-04-16 (Christophe Dunand) |
| Last sequence changes | 2009-04-16 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Christophe Dunand |
| Last annotation changes | 2023-01-09 (Christophe Dunand) |
Peroxidase information: ZmPrx70(Zm00001d022458 / GRMZM2G025441 / pmPOX2b / PRX70 / Peroxidase 70)
| Name | ZmPrx70(Zm00001d022458 / GRMZM2G025441 / pmPOX2b / PRX70 / Peroxidase 70) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx064] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Monocotyledons Poaceae Zea | ||||||||||||||||||||||||||||||||||||
| Organism | Zea mays [TaxId: 4577 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | Roots |
||||||||||||||||||||||||||||||||||||
| Inducer | Drought |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
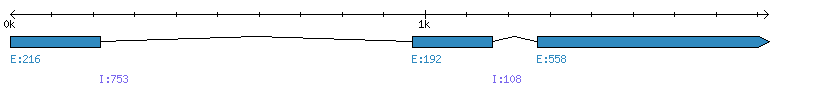
| Gene structure Fichier |
Exons►
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references ZmPrx70(Zm00001d022458 / GRMZM2G025441 / pmPOX2b / PRX70 / Peroxidase 70)
| Literature | Mika,A. and Luthje,S. Production of active oxygen species and sequence analysis of plasma membrane-bound peroxidases from maize (Zea mays L.) roots. |
|---|---|
| Protein ref. | UniProtKB: A5H452 |
| DNA ref. | GenBank: AC210153.3 (99674..101500) CG186903 (99674..101497) |
| mRNA ref. | GenBank: EF059717 |
| Cluster/Prediction ref. | UniGene: Zm.10660 |
Protein sequence: ZmPrx70(Zm00001d022458 / GRMZM2G025441 / pmPOX2b / PRX70 / Peroxidase 70)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (Chromo 7, introns 1 and 2), 1 mRNA and 10 ESTs. Incorrect prediction from Phytozome (Exon 1 is not predicted). |
||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
