Entry information : ZmAPx[P]07(Zm00001d054096 / GRMZM5G877989)
| Entry ID | 8006 |
|---|---|
| Creation | 2011-02-18 (Christophe Dunand) |
| Last sequence changes | 2011-02-18 (Christophe Dunand) |
| Sequence status | theoretical translation / pseudogene |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2022-12-29 (Christophe Dunand) |
Peroxidase information: ZmAPx[P]07(Zm00001d054096 / GRMZM5G877989)
| Name | ZmAPx[P]07(Zm00001d054096 / GRMZM5G877989) | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Ascorbate peroxidase [Orthogroup: APx001]* | ||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Monocotyledons Poaceae Zea | ||||||||||||||||||||||||||||||||||||||||||||||||
| Organism | Zea mays [TaxId: 4577 ] | ||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
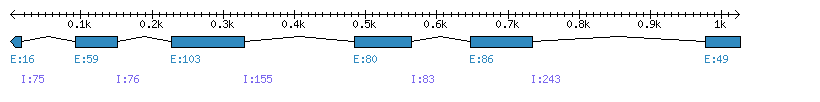
|
||||||||||||||||||||||||||||||||||||||||||||||||
Literature and cross-references ZmAPx[P]07(Zm00001d054096 / GRMZM5G877989)
Protein sequence: ZmAPx[P]07(Zm00001d054096 / GRMZM5G877989)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Pseudogene (chromo 4). Large 5' end is missing. Incorrect prediction from Phytozome. | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
