Entry information : GmPrx105 (Glyma01g40870.1)
| Entry ID | 8935 |
|---|---|
| Creation | 2011-07-04 (Qiang Li) |
| Last sequence changes | 2011-10-03 (Qiang Li) |
| Sequence status | complete |
| Reviewer | Christophe Dunand |
| Last annotation changes | 2011-12-13 (Qiang Li) |
Peroxidase information: GmPrx105 (Glyma01g40870.1)
| Name (synonym) | GmPrx105 (Glyma01g40870.1) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx043] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Fabaceae Glycine | ||||||||||||||||||||||||||||||||||||
| Organism | Glycine max (soybean) [TaxId: 3847 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
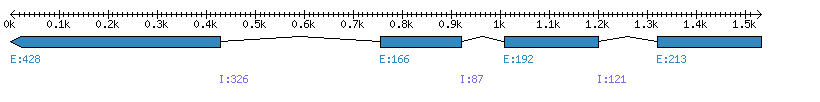
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references GmPrx105 (Glyma01g40870.1)
| DNA ref. | Phytozome 12: Gm01 (52518582..52517050) |
|---|
Protein sequence: GmPrx105 (Glyma01g40870.1)
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic. Incorrect prediction from Phytozome (5' end is missing). No EST. | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
