Entry information : CgraGPx01
| Entry ID | 9681 |
|---|---|
| Creation | 2011-11-10 (Passaia Gisèle) |
| Last sequence changes | 2011-11-14 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2016-03-08 (Christophe Dunand) |
Peroxidase information: CgraGPx01
| Name | CgraGPx01 | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Fungi-Bacteria glutathione peroxidase [Orthogroup: Gpx3001] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Fungi Ascomycota Sordariomycetes Glomerellaceae Glomerella | ||||||||||||||||||||||||||||||||||||
| Organism | Colletotrichum graminicola (Glomerella graminicola) [TaxId: 31870 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | N/D |
||||||||||||||||||||||||||||||||||||
| Inducer | N/D |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
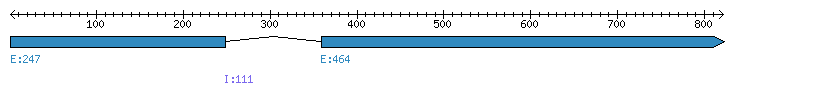
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references CgraGPx01
| DNA ref. | GenBank: ACOD01000058.1 (411397..412218) |
|---|---|
| EST ref. | GenBank: FE741948.1 [5' end] FE742228.1 [3' end] |
Protein sequence: CgraGPx01
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (Supercontig 9, 1 intron) and 2 ESTs. Strain="M1.001" | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
