Entry information : SbPrx26 (Sobic.002G003500.1[2.1] / SB02G000500[1.4])
| Entry ID | 985 |
|---|---|
| Creation | 2005-12-19 (Christophe Dunand) |
| Last sequence changes | 2015-02-04 (Christophe Dunand) |
| Sequence status | complete |
| Reviewer | Not yet reviewed |
| Last annotation changes | 2015-02-04 (Christophe Dunand) |
Peroxidase information: SbPrx26 (Sobic.002G003500.1[2.1] / SB02G000500[1.4])
| Name (synonym) | SbPrx26 (Sobic.002G003500.1[2.1] / SB02G000500[1.4]) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Class III peroxidase [Orthogroup: Prx058] | ||||||||||||||||||||||||||||||||||||
| Taxonomy | Eukaryota Viridiplantae Streptophyta Monocotyledons Poaceae Sorghum | ||||||||||||||||||||||||||||||||||||
| Organism | Sorghum bicolor [TaxId: 4558 ] | ||||||||||||||||||||||||||||||||||||
| Cellular localisation | N/D |
||||||||||||||||||||||||||||||||||||
| Tissue type | Seedlings |
||||||||||||||||||||||||||||||||||||
| Inducer | Phosphate starvation |
||||||||||||||||||||||||||||||||||||
| Repressor | N/D |
||||||||||||||||||||||||||||||||||||
| Best BLASTp hits |
|
||||||||||||||||||||||||||||||||||||
| Gene structure Fichier |
Exons►
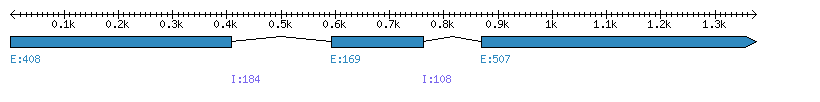
|
||||||||||||||||||||||||||||||||||||
Literature and cross-references SbPrx26 (Sobic.002G003500.1[2.1] / SB02G000500[1.4])
| Literature | Cordonnier-Pratt,M.-M. et al., unpublished |
|---|---|
| DNA ref. | Phytozome 12: chromosome_2 (451565..452828) |
| Cluster/Prediction ref. | PlantGDB: SbGSStuc11-12-04.1313.1 |
Protein sequence: SbPrx26 (Sobic.002G003500.1[2.1] / SB02G000500[1.4])
| Sequence Properties first value : protein second value (mature protein) |
|
||||||||||||
| Sequence Send to BLAST Send to Peroxiscan |
|
||||||||||||
| Remarks | Complete sequence from genomic (chromo 2, introns 2 and 3) and 2 ESTs. Frame shift between "VSPRL" and "YNFGG" (probably missing "t"). Incorrect prediction from Phytozome. Containing motif 'SVDC'. Missing 'Cys'. | ||||||||||||
| DNA ► Send to BLAST |
|
||||||||||||
| CDS► Send to BLAST |
|
||||||||||||
