Ever since 2004, the PeroxiBase was a database dedicated to most of the peroxidase superfamilies.
Since 2017, the RedoxiBase centralizes peroxidases and other oxido-reductase together with the compilation of the information concerning putative
functions and transcriptional regulation.
The RedoxiBase also provides annotated and manually checked sequences.
With the global overview of several Oxido-reductase classes phylogeneticaly related, we expect to better understand the evolution of the superfamilies
among living organisms.
Quality problems related to automatic annotation of genomes, assembly and clustering of ESTs are still relevant. At our level, they are especially important as the number of genome sequencing projects has exploded in recent years. In addition to the NGS technologies, we are still faced with the build quality for large multigene families.
To address these quality issues, we continue to produce an expert annotation combined with a semi automatic annotation of genome, proteome and NGS data. Using the highly conserved motifs of each peroxidases class, manual annotation and editing allow identifying clearly the correct sequences even in low quality sequences, updating the existing sequences and increasing the number of annotated sequences.
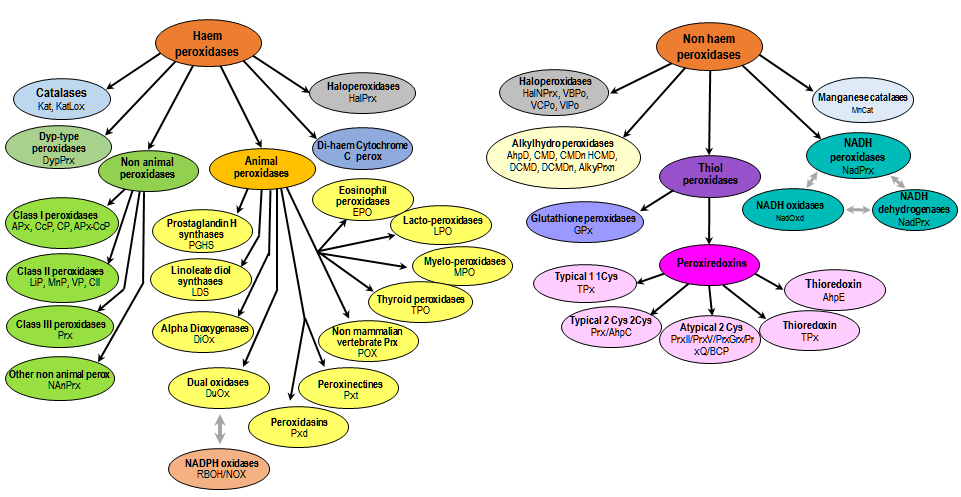
Grey arrows link two different protein classes but sharing common domains.
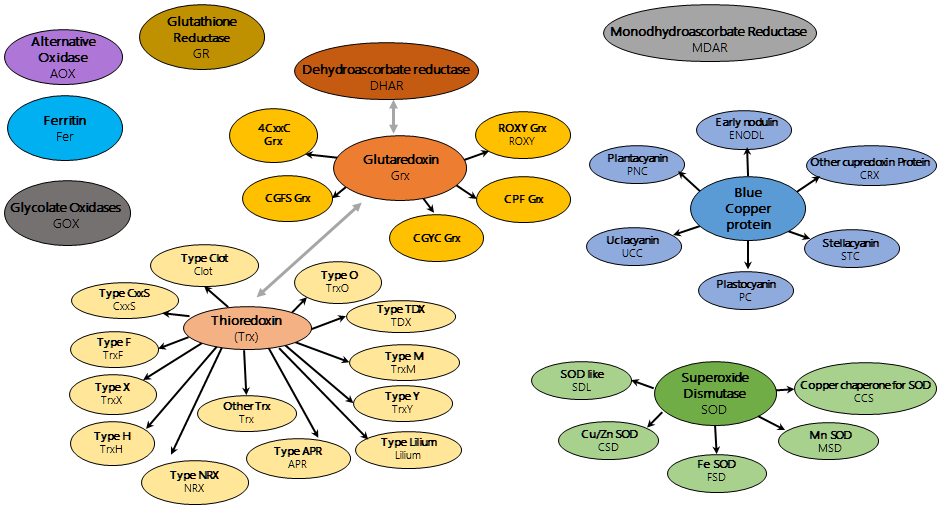
Grey arrows link two different protein classes but sharing common domains.
