About the RedoxiBase
Peroxidases and other oxido-reductase are enzymes able to catalyze reduction of hydrogen peroxide and oxidize various substrates. This biochemical function confers them a role in many different and important biological processes such as defence mechanism, immune response, pathogeny. These proteins are present in all kingdoms and are members of multigenic families (from 2 to more then hundred isoforms).
Using the highly conserved motifs of each peroxidases and other oxido-reductases classes, manual annotation and editing allow identifying clearly the correct sequences even in low quality sequences, updating the existing sequences and increasing the number of annotated sequences.
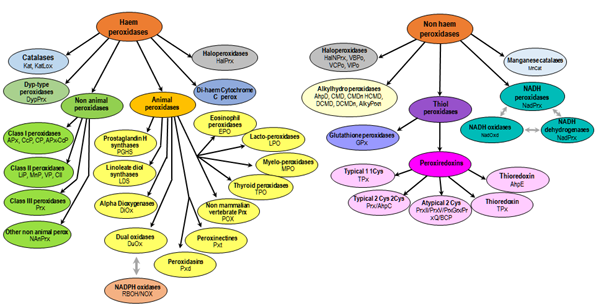
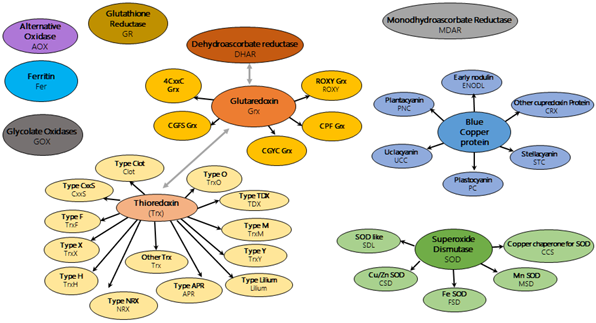
The goal of the RedoxiBase is to centralize most of the oxido-reductase superfamilies encoding sequences (see the schema below), to follow the evolution of the different families among living organisms and to compile the information concerning putative functions and transcriptional regulation.